Research
MSM/RD: Multiscale molecular kinetics by coupling Markov state models with particle-based reaction-diffusion simulations
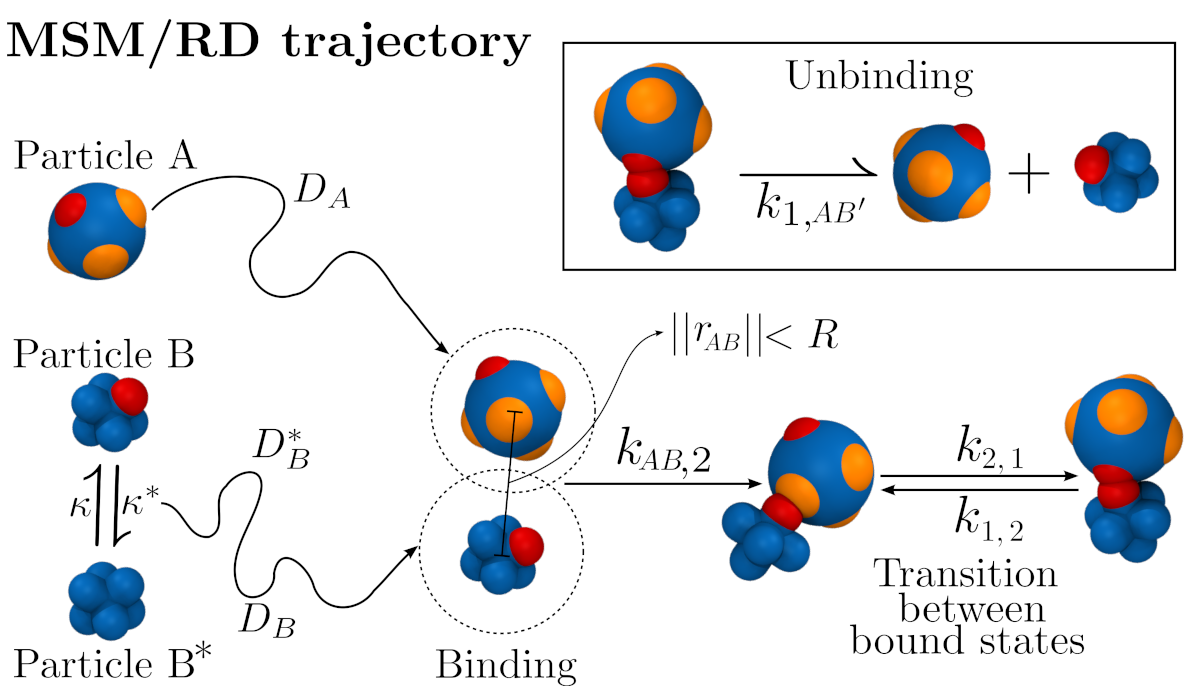
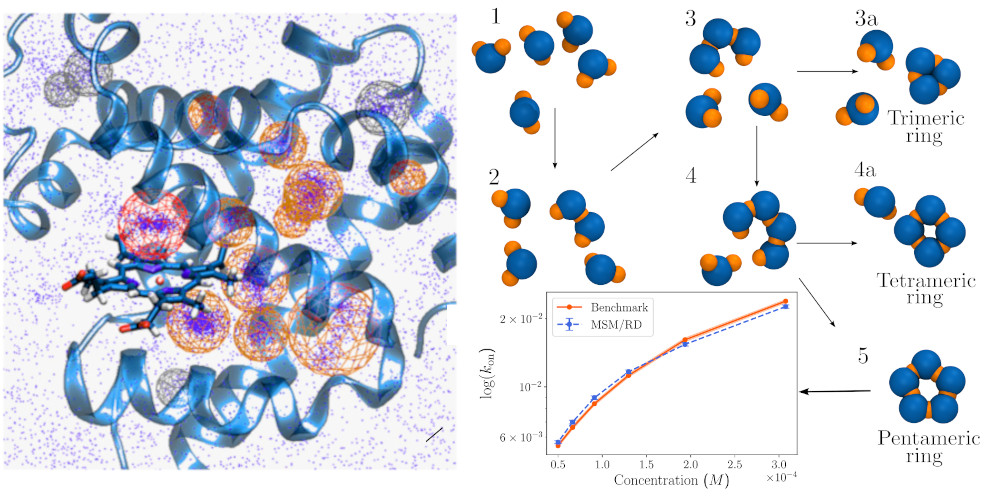
Molecular dynamics (MD) simulations can model the interactions between macromolecules with high spatiotemporal resolution but at a high computational cost. By combining MD with Markov state models (MSMs), it is possible to obtain long-timescale behavior of small to intermediate biomolecules and complexes. On the other hand, to model the interactions of many molecules at large lengthscales, particle-based reaction-diffusion (RD) simulations are more suitable but come at the price of sacrificing molecular detail. This project is born with the desire to couple MSMs and RD simulations (MSM/RD) to obtain the best of both worlds by efficiently producing simulations at large time- and lengthscales, while still conserving molecular resolution. The essence of the method is depicted in the image above. The images below show applications to protein-ligand systems (binding of CO to myoglobin) and mutiparticle extensions (pentameric ring protein formation).
Related publications:
-
M. J. del Razo, M. Dibak, C. Schütte, and F. Noé (2021) Multiscale molecular kinetics by coupling Markov state models and reaction-diffusion dynamics. [arXiv] [github]
-
M. Dibak, M. J. del Razo, D. De Sancho, C. Schütte and F.Noé (2018) MSM/RD: Coupling Markov state models of molecular kinetics with reaction-diffusion simulations. The Journal of Chemical Physics 148, 214107 [arXiv]
Multiscale reaction-diffusion processes in open settings: theory and simulation
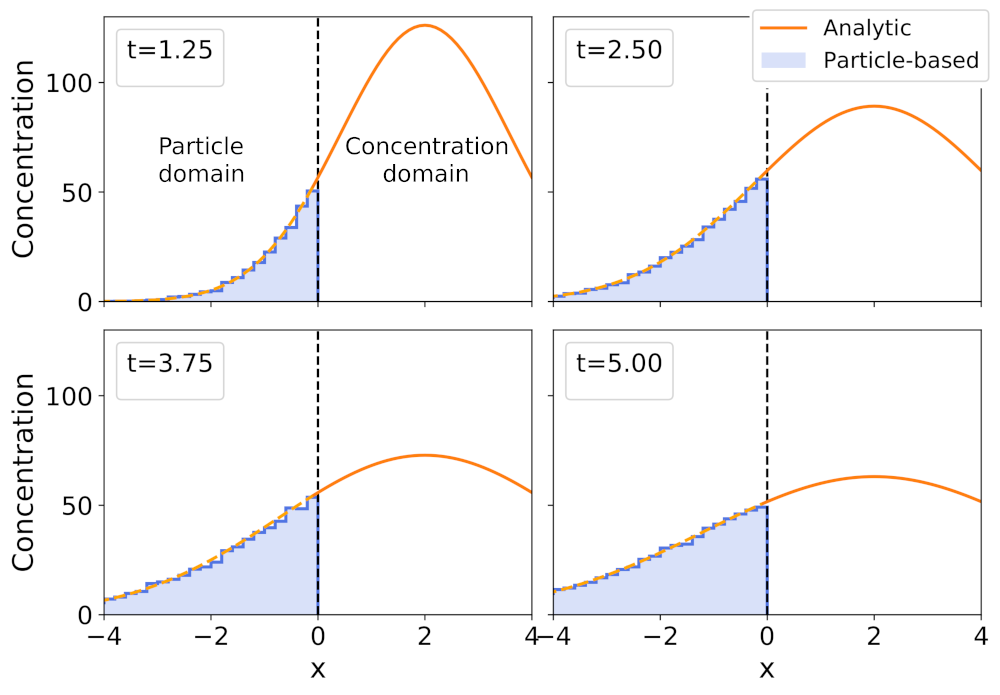
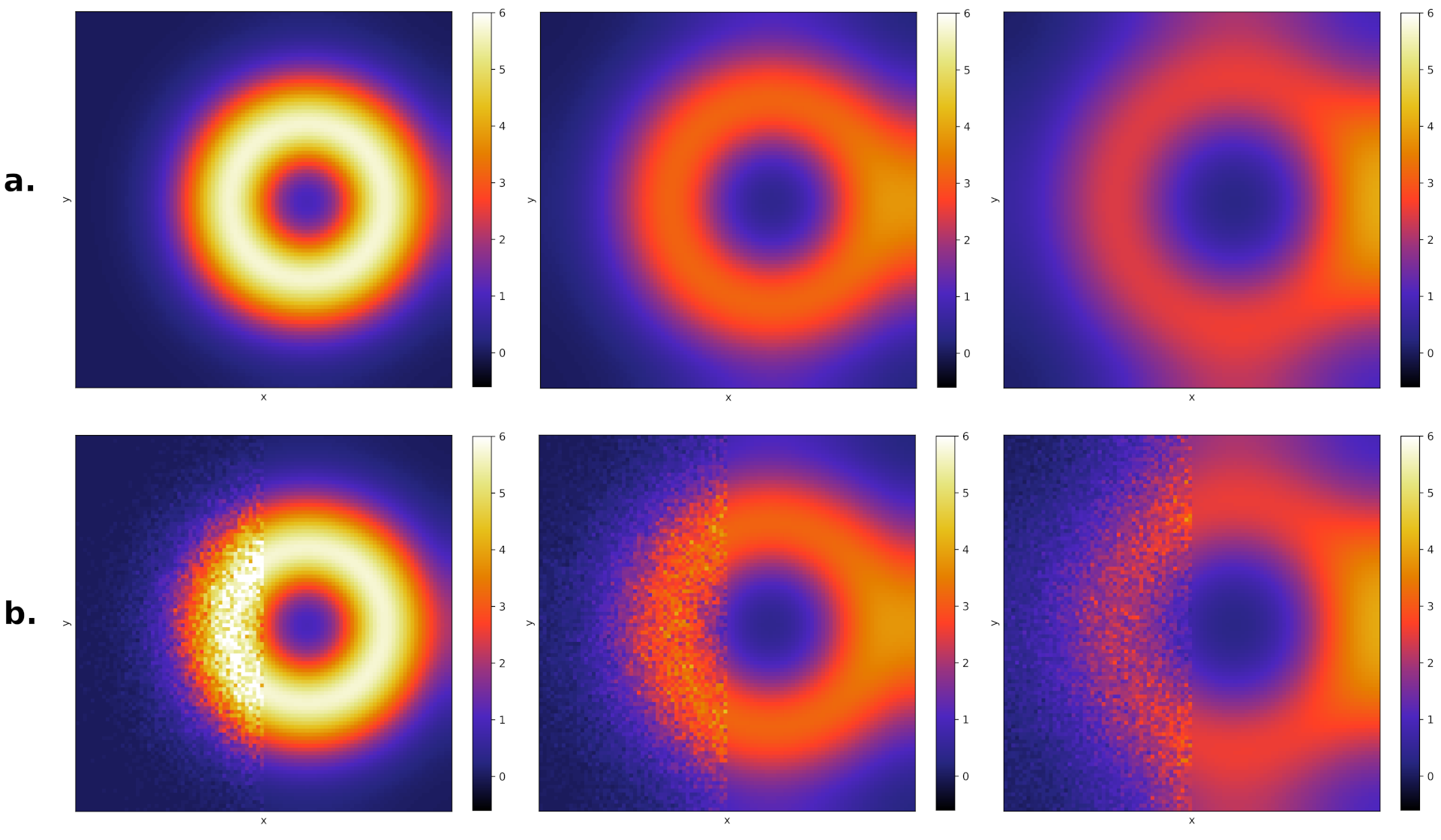
Open biochemical systems of interacting molecules are ubiquitous in life-related processes. However, established computational methodologies, like molecular dynamics, are still mostly constrained to closed systems and timescales too small to be relevant for life processes. Alternatively, particle-based reaction-diffusion models are currently the most accurate and computationally feasible approach at these scales. Their efficiency lies in modeling entire molecules as particles that can diffuse and interact with each other. In this work, we develop modeling and numerical schemes for particle-based reaction-diffusion in an open setting, where the reservoirs are mediated by reaction-diffusion PDEs. This allows modeling open biochemical systems in contact with reservoirs that are time-dependent and spatially inhomogeneous, as in many relevant real-world applications.
In the image above, we show the coupling results for a one-dimensional diffusion process. The right half corresponds to the reservoir modeled by a PDE, and the left half corresponds to a histogram resulting from averaging over several particle-based simulations. In the image to the left, we do the same for a two-dimensional Lotka Volterra model (predator-prey). We show only the solution for the concentration of prey. The top row (a.) corresponds to the full PDE solution at three times. The bottom row (b.) corresponds to the hybrid simulation of the same three times. Once again the right half is the PDE solution, and the left half is the averaged solution over several particle-based simulations. The solutions are very accurate because of two reasons: we derived mathematical results that relate the particle-based simulation parameters with the PDE parameters, and we develop a computational framework to consistently couple the different scales.
Related publications:
-
M. Kostré, C. Schütte, F. Noé and M. J. del Razo (2020) Coupling particle-based reaction-diffusion simulations with reservoirs mediated by reaction-diffusion PDEs. [arXiv]
- M. J. del Razo, H. Qian and F. Noé (2018) Grand canonical diffusion-influenced reactions: a stochastic theory with applications to multiscale reaction-diffusion simulations. The Journal of Chemical Physics 149, 044102 [arXiv]
- M. J. del Razo and H. Qian (2016) A discrete stochastic formulation for reversible bimolecular reactions via diffusion encounter. Communications in Mathematical Sciences, Vol. 14, No. 6, pp. 1741-1772. [arXiv]
Previous projects
Under construction …